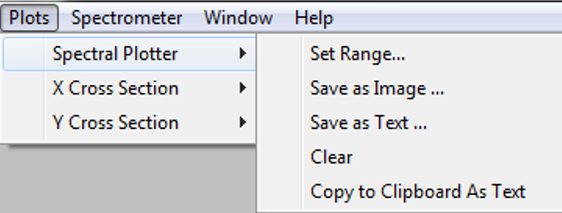
6. Advanced Data Analysis 1: General¶
Spectronon’s data visualization and analysis tools are presented in this section, following the structure of the main menu.
6.7. Datacubes and Header Files¶
Datacubes collected with Resonon imaging spectrometers require two files, one that contains the hyperspectral data, and also a header file that contains important information about the data. Hyperspectral data can be accessed with many tools, for example, Matlab and ENVI, and is compatible with a variety of languages including: Python, C++, Fortran.
6.7.1. Hyperspectral data files¶
ENVI-compatible hyperspectral data can be stored in the following three formats:
Band-Sequential In Band-Sequential (BSQ) format each line of the data is followed by the next line in the same spectral band. Hyperspectral data stored in BSQ format will end with .bsq.
Band-Interleaved-by-Line Data saved in Band-Interleaved-by-Line (BIL) format have the first line of the first band followed by the first line of the second band, followed by the first line of the third band, and so forth. This pattern is repeated for subsequent lines. Hyperspectral data stored in BIL format will end with .bil.
Band-Interleaved-by-Pixel Band-Interleaved-by-Pixel (BIP) data have the first pixel for all bands in sequential order, followed by the second pixel for all bands, and so forth for all pixels. Hyperspectral data stored in BIP format will end with .bip.
Resonon imaging spectrometers will save hyperspectral data in BSQ, BIL, or BIP formats, and Spectronon software will open hyperspectral data in all of these formats.
6.7.2. Hyperspectral data header files¶
Header files for hyperspectral data have the same name as the BSQ, BIL, or BIP files followed by .hdr. The following discussion provides an overview of header files.
Header files can be opened and edited using WordPad. The following list describes some, but not all, of header file commands. Bold indicates the command, quantities in (brackets) are acceptable arguments for the commands, and statements in [square brackets] are additional explanation. Note that the order of these commands can be different from file to file.
ENVI [All ENVI-compatible header files begin with this]
interleave = (bil, bip, bsq) [Tells the software the hyperspectral data format]
data type = (4, 12) [4 is 4-byte floating point; 12 is 2-byte unsigned ; other data types are also supported, but these are common]
lines = (###) [The number of lines in the hyperspectral data where ### is a number]
samples = (###) [The number of samples in the hyperspectral data where ### is a number. This is typically the number of cross-track spatial pixels of the hyperspectral imager.]
bands = (###) [The number of bands in the hyperspectral data where ### is a number]
bit depth = (12, 14) [The bit depth of the hyperspectral data]
shutter = (###) [The shutter time in units of msec where ### is a number. This is mostly for reference, but it may be used by certain plugins]
gain = (#.#) [The camera gain in dB where #.# a number. This is mostly for reference, but it may be used by certain plugins]
framerate = (####) [The framerate at which the data were recorded in frames/sec where ### is a number. This is mostly for reference, but it may be used by certain plugins]
reflectance scale factor = (4095, 1023, 1) [If you divide the data by this factor, the data will scale from 0 to 1, typically for reflectance data]
byte order = (0) [byte order is the ordering of byte significance (http://en.wikipedia.org/wiki/Endianness) Data for Spectronon are LSF or Little Endian which means the least significant bytes come first. Spectronon will not properly open BSF or Big Endian files]
header offset = (0) [This is the number of bytes at the beginning of the binary file that should be ignored].
wavelengths = (###, ###, ###, … ) [List of the spectral band centers in units of nm]
rotation = ((#,#), (#,#), (#,#), (#,#)) [This is a default orientation of the view of a datacube in Spectronon. This is a Spectronon only header option].
label = (name) [The label is used by Spectronon to give a human readable alternative to the file name of a file. If omitted, Spectronon will use the file name
timestamp = (day & time of datacube) [If available, this is when the datacube was recorded. The timestamp is often found on airborne datacubes, and datacubes where a script entered this information explicitly into the header]
The following are for spec.hdr files only, headers for spectrum. Spectrum files are ENVI compatible because they are 1x1 datacubes, but Spectronon adds in these data for its own use.
original cube file = (name of original cube) [This is a Spectronon-only header item for recording the history of a generated spectrum. In the case of a derived datacube, you will see a “history” header value that shows the way this cube was generated]
pointlist = (long list of values) [This is the set of points from the original cube that were averaged together to make this spectrum if the spectrum is a mean]
label color = (#FF00FF) [This is the color used to plot the spectrum]
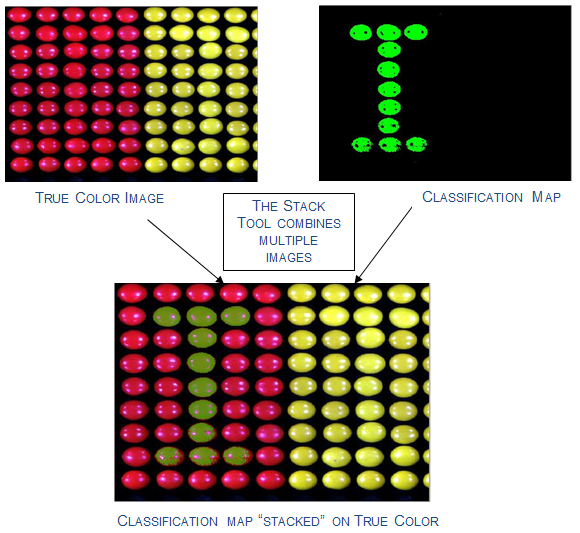

 or
or  ) or by loading a previously saved
spectrum or cube.
) or by loading a previously saved
spectrum or cube.
 , or
use one of the ROI tools
, or
use one of the ROI tools 
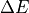 values for each pixel as compared to a standard set by
the user.
values for each pixel as compared to a standard set by
the user.

 ). For all of the
following options, you must first select a Region of Interest (ROI).
). For all of the
following options, you must first select a Region of Interest (ROI).